| Version 125 (modified by dgoll, 7 years ago) (diff) |
|---|
ORCHIDEE-CN-P (former branch ORCHIDEE-CNP)
This page describes the phosphorus cycle in ORCHIDEE-CN-P. It is based on ORCHIDEE-CN, which was extended and corrected as described here: https://forge.ipsl.jussieu.fr/orchidee/wiki/Branches/MergeOCN/Goll as well as additional non-documented bugfixes to avoid negative pools due to machine precisions as well as bugs.
The phosphorus cycle is an adaptation of the the model described by http://www.biogeosciences.net/9/3547/2012/bg-9-3547-2012.html . Nonetheless, the complexity was substantially increased due to the more detailed representation of the C and N cycle in ORCHIDEE compared to JSBACH.
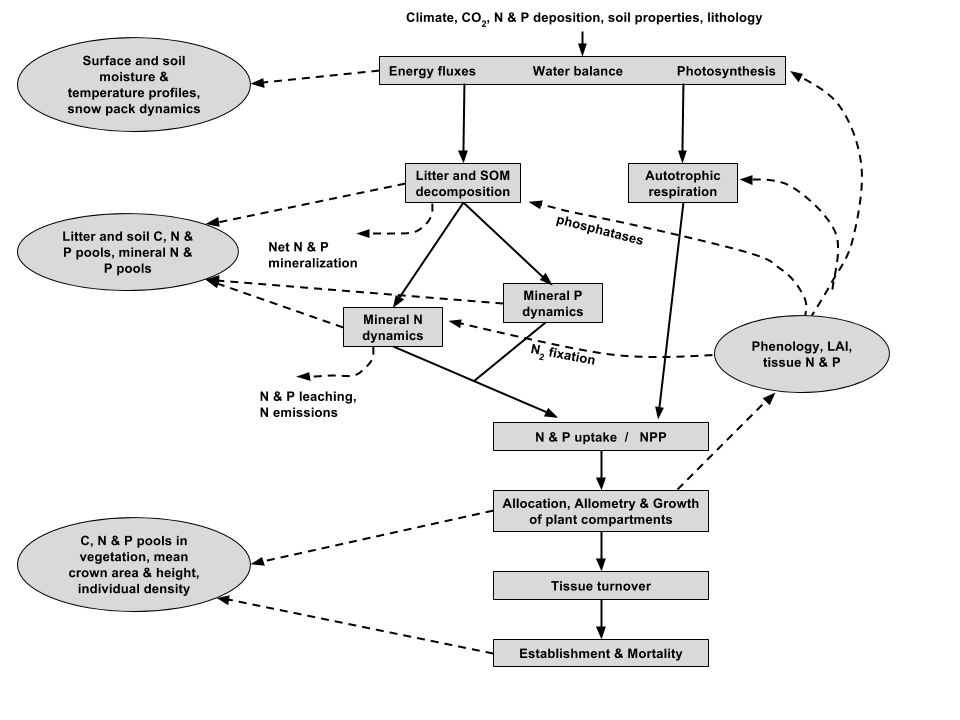 Schematic representation of the key processes represented in ORCHIDEE-CNP
Schematic representation of the key processes represented in ORCHIDEE-CNP
2. Technical modification to the nitrogen code
I modified the code a bit to avoid problems and redundant code. More information here: https://forge.ipsl.jussieu.fr/orchidee/wiki/DevelopmentActivities/ORCHIDEE-CNP/TechnicalMods
3. Conceptual modifications to the nitrogen cycle
3.1 soil mineral N concentration in soil solution
Following Smith et al (2014), I introduced the use of the maximum water holding capacity of soils (max_var_eau) to approximate pore space which to derive the average soil mineral N concentration in solution. The use of the actual water volume can not be recommended as we this would lead to high N concentration in soil water when soil water is very low. As we do not account for the inhibition of replenishment of mineral N in the soil solution around roots when soil water is scarce.
3.2 soil mineral N dynamics
ORCHIDEE-CN performs poorly in respect to soil mineral N dynamics. This is due to two reason. First, ORCHIDEE-CN is based on Zaehle & Friend (2010) and does not incorporate the modification to the DNDC formulations which avoid instabilities when run on global scale (Zaehle et al., 2011). Second, most of the modifications in ORCHIDEE-CN I assume are not tested, as most of the environmental scaling function "explode" with the parameter set used, or are not sound imo.
I step by step replaced all modification by using the formulation in Zaehle et al. (2011) (http://://www.nature.com/ngeo/journal/v4/n9/extref/ngeo1207-s1.pdf), testing for the size of the mineral N stocks as well as ensuring all scaling function work well in an global environmental space.
Final mineral N dynamics The mineral N dynamics in ORCHIDEE-CN-P are now exactly as described in Zaehle et al. (2011), except that I did not take over the rain drainage formulation as we use an improved hydrological scheme compared to Zaehle et al. (2011) which is able to simulate the rain drainage as a result of more soil layers (11 layers).
The mineral N stocks of the test sites are all in realistic ranges. Pattern correlation test with N15 derived fraction of denitrification show poor correlation due to high NOx emissions. This needs to be corrected in the near future.
3.3 Biological N2 fixation (BNF)
3.3.1 Modified Cleveland et al. (1999) scheme
We introduce a module which computes BNF as a function of NPP, tissue C:N, and tissue & N:P. This approach is based on Cleveland et al (1999), Thornton et al. (2007), and Goll et al. (2012). When using the analytical spinup the BNF must be read in from a file to speed up the spin up. The file should contain the reference BNF rates for the respective climatic conditions. Such files are currently missing, but should be generated as soon as the model is ready.
3.3.2 Biological N2 fixation (BNF) from Yingping
The Cleveland scheme is heavily criticized due to good reason. Yingping Wang will provide maps with annual rates of BNF computed based more or less on Wang et al. (2007) for the historical period. To do so, Yingping needs TRENDY style simulations with a calibrated model.After publication of his model, he will share the code with us & we can compute BNF dynamically in ORCHIDEE.
4. Re-calibration of the carbon cycle
The calibration of ORCHIDEE-CAN of the carbon cycle in combination with the Vcmax being derived from leaf nitrogen content, leads to unrealistic C cycle (CUE, biomass stocks, biomass turnover, etc.). Using as a guide rough ranges of observation-based estimates of aboveground biomass, LAI, CUE, and biomass turnover, we manipulated parameters of the pipe model (TAU_SAP, KLATOSA_MIN, K_LATOSA_MAX) as well as the inversion of the rate constant for tree turnover RESIDENCE_TIME and the factor relating maintenance respiration to tissue N, COEFF_MAINT.
The main issue with the previous calibration (Naudts et al GMD) is, that FLUXNET GPP & energy fluxes as well as LAI estimates are poor constraints on biomass and biomass turnover when calibrating the ORCHIDEE-CAN parameter. The clear cuts in the management simulations in the GMD hide the potential biomass stocks. Even the biomass stocks estimates which exclude managed forest are smalled than the ones in ORCHIDEE.
The range of K_LATOSA prescribed makes things tricky as total biomass is very sensitive to the actual values of K_LATOSA. The dynamics of K_LATOSA were several times revised and it is not clear what is going on.
I derived a set of parameters which result in kind of okay LAI, total biomass, biomass turnover and CUE. Still some PFTs behave not really realistic, like DBF PFTs.
ISSUE with my calibration: To facilitate the calibration I derive maintenance respiration only from leaf N and not from sapwood N too. This allows to manipulate total biomass via K_LATOSA without affecting the CUE. It would be better to have it coupled as this adds a constraint to K_LATOSA. Best would be to include even more constraints on K_LATOSA for example hydrological constraints (like the stuff Emilie develops).
link to the results: https://docs.google.com/presentation/d/19IohhIXFAYZQRzbSp616-C7dRdsClQYlu9EQSqHFN7U/edit?usp=sharing
5. New input files
You can find information on the new input files needed for the P cycle here: https://forge.ipsl.jussieu.fr/orchidee/wiki/DevelopmentActivities/ORCHIDEE-CNP/newInput
6. Notes regarding Vcmax and leaf nutrients
https://forge.ipsl.jussieu.fr/orchidee/wiki/DevelopmentActivities/ORCHIDEE-CNP/Vcmax
7. unresolved issues (ideas for future developments)
There are still yet unresolved issue with the model. They are listed here: https://forge.ipsl.jussieu.fr/orchidee/wiki/DevelopmentActivities/ORCHIDEE-CNP/issues
8. Howto install, compile & run the model
You can find information on how to setup the simulations here: https://forge.ipsl.jussieu.fr/orchidee/wiki/DevelopmentActivities/ORCHIDEE-CNP/howtoUse
9. Some results
You can find some basic plots of a outdated model version here: https://forge.ipsl.jussieu.fr/orchidee/wiki/DevelopmentActivities/ORCHIDEE-CNP/results
Attachments (11)
- missingPoints.png (20.5 KB) - added by dgoll 8 years ago.
-
USDA_map.png
(82.4 KB) -
added by dgoll 8 years ago.
The input file with the USDA soil orders in alphabetical order
-
USDA_map.2.png
(76.8 KB) -
added by dgoll 8 years ago.
input file with USDA soil orders in alphabetical order
-
USDA_map.3.png
(76.0 KB) -
added by dgoll 8 years ago.
The input file with the USDA soil orders in alphabetical order
-
lith_dom_map.png
(96.2 KB) -
added by dgoll 8 years ago.
dominant lithology
-
soilshield_map.png
(83.9 KB) -
added by dgoll 8 years ago.
soil shielding factor
-
ORC-CNP.png
(69.6 KB) -
added by dgoll 8 years ago.
Schematic representation of the key processes represented in ORCHIDEE-CNP
-
CNP_longterm.png
(46.6 KB) -
added by dgoll 8 years ago.
In red original formulation, in green the 3yr average. Shown is the C, the N and the CN of slow soil pool.
- BurnLabile.png (106.3 KB) - added by dgoll 8 years ago.
- Zoom.png (57.2 KB) - added by dgoll 8 years ago.
-
GPP_Hai.png
(48.2 KB) -
added by dgoll 8 years ago.
average annual cycle of DE-Hai site w & w/o N-limitation / w & w/o burning
Download all attachments as: .zip
