| Version 72 (modified by dgoll, 8 years ago) (diff) |
|---|
ORCHIDEE-CN-P (former branch ORCHIDEE-CNP)
This page describes the work done to include the phosphorus cycle into ORCHIDEE. It is based on MERGE-OCN, which was extended and corrected as described here: https://forge.ipsl.jussieu.fr/orchidee/wiki/Branches/MergeOCN/Goll as well as a recruitment routine by S. Peng (ask him if you need infos) The phosphorus cycle is an adaptation of the the model described by http://www.biogeosciences.net/9/3547/2012/bg-9-3547-2012.html
modularisation of code used by nitrogen and phosphorus routines
There are many aspects which the nutrient cycles have in common, for example root uptake kinetics, stoichiometric considerations, etc. To avoid the risks of introducing inconsistencies and redundant code, I started to write subroutines which are listed in the following. All subroutines are in stomate_phosphorus.f90.
1. root_conductivity
This subroutine calculates the uptake capacity of N & P per mass root according to the uptake kinetics of ammonia and nitrate used in OCN (Zaehle & Friend, 2010).
2. f_XY_plant
This subroutine calculates the scaling functions based on the stoichiometric ratio of labile plant tissue (reserve, labile and leaf) which are used to scale root uptake, biological N2 fixation, and biochemical mineralization. It currently supports scaling functions based on the P-to-N ratio, N-to-C ratio and P-to-C ratio. All other combinations will cause a 'STOP', but could be easily implemented if needed.
New input files
USDA soil orders
We use the USDA soil order map of Sun et al. (in revision) with the dominant soil order per pixel. The gap filling of the missing values was done like this (see script /home/users/dgoll/ORC_data/USDA_soilorders/try_2_fill/fill_deserts.ksh):
We use the soiltext class 6 for cold desert from the input file "soils_param.nc" to fill missing values in these region with Gelisols (5). After we set cold desert, we use the level 1 of the maxvegetfrac in the input file "PFTmap.nc" to fill missing values of the remaining desert to Aridisols (2). Hereby, we use a threshold of 0.8 for identifying deserts in maxvegetfrac.
In case of the remaining missing points, mostly coast line points, ORCHIDEE will assign the value of the nearest neighbour in the routine "get_soil_orders" in src_stomate_io.f90.
 Figure 1: USDA soil orders: Alfisols(1), Andisols(2), Aridisols(3), Entisols(4), Gelisols(5), Histosols(6), Inceptisols(7), Mollisols(8), Oxisols(9), Spodosols(10),Ultisols(11), Vertisols(12)
Figure 1: USDA soil orders: Alfisols(1), Andisols(2), Aridisols(3), Entisols(4), Gelisols(5), Histosols(6), Inceptisols(7), Mollisols(8), Oxisols(9), Spodosols(10),Ultisols(11), Vertisols(12)
GliM lithology
We use the GliM lithology map by Hartmann & Moosdorf (2012). We account for subgrid scale heterogeneity by reading the fractional coverage of each of the 16 classes.
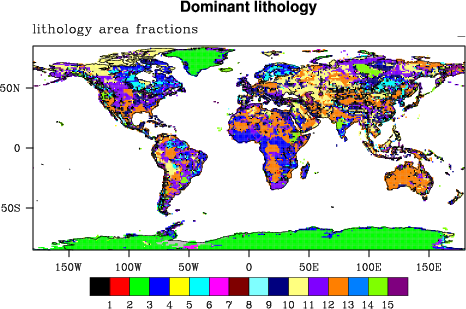 Figure 2: GliM lithology: the dominant type with a fractional cover of >0.51 ! alphabetical sequence of lithological classes:
! ! 0 - no dominant type
! ! 1 - evaporites
! ! 2 - ice & glaciers
! ! 3 - metamorphics
! ! 4 - no data
! ! 5 - acid plutonic rocks
! ! 6 - basic plutonic rocks
! ! 7 - intermediat plutonic rocks
! ! 8 - pyroclastics
! ! 9 - carbonate sedimentary rocks
! ! 10 - mixed sedimentary rocks
! ! 11 - siliciclastic sedimentary rocks
! ! 12 - unconsolidated sediments
! ! 13 - acid volcanic rocks
! ! 14 - basic volcanic rocks
! ! 15 - intermediate volcanic rocks
! ! 16 - water bodies
Figure 2: GliM lithology: the dominant type with a fractional cover of >0.51 ! alphabetical sequence of lithological classes:
! ! 0 - no dominant type
! ! 1 - evaporites
! ! 2 - ice & glaciers
! ! 3 - metamorphics
! ! 4 - no data
! ! 5 - acid plutonic rocks
! ! 6 - basic plutonic rocks
! ! 7 - intermediat plutonic rocks
! ! 8 - pyroclastics
! ! 9 - carbonate sedimentary rocks
! ! 10 - mixed sedimentary rocks
! ! 11 - siliciclastic sedimentary rocks
! ! 12 - unconsolidated sediments
! ! 13 - acid volcanic rocks
! ! 14 - basic volcanic rocks
! ! 15 - intermediate volcanic rocks
! ! 16 - water bodies
SECTIONS
ISSUES
Reading in N inputs
The variables storing the N inputs in the restart file are all undef. Not quite sure why. I set them all to zero.
Soil type inconsistency in ORCHIDEE-CN-P
The phosphorus model needs soil type specific parameters. Therefore, a new input routine is going to be written which reads in USDA soil orders. According to the soil order the parameter values are set for each grid box.
The problem is that ORCHIDEE already reads in fields of soil texture classes which were originally derived from soil orders. According to the soil texture classes, parameter related to soil hydrology are set. As the original data cannot be found anymore, ORCHIDEE will run with two independent sets of soil order specific parameters. Thereby inconsistencies between the phosphorus related parameters and the the hydrology related parameters can be introduced. The practice of reading in soil texture classes instead of soil orders is also problematic in respect of future developments and I suggest to change that.
HOWTO install/compile/run ORCHIDEE-CN-P for offline use with libIGCM.
This howto is based on the information which can be found in the ORCHIDEE wiki regarding the trunk version of ORCHIDEE. Please check the wiki for information regarding the respective commands listed here.
1. install libIGCM
1.1 where to install
You should install the model NOT in the home directory; it is even too small to handle diagnostics like "out_execution" or "out_orchidee". You can use , for example, your work directory: curie:
/ccc/work/cont003/dsm/<userID>
or project storage; here example for the IMBALANCE-P project on obelix:
/home/surface3/dgoll
1.2 install libIGCM & trunk ORCHIDEE
svn co http://forge.ipsl.jussieu.fr/igcmg/svn/modipsl/trunk modipsl cd modipsl/util ./model ORCHIDEE_trunk
2. Exchange the trunk ORCHIDEE with ORCHIDEE-CN-P
cd ../modeles rm -fr ORCHIDEE svn co svn://forge.ipsl.jussieu.fr/orchidee/branches/ORCHIDEE-CN-P ORCHIDEE cd ../util ; ./ins_make
3. Compile ORCHIDEE
adjust IOIPSL to be able to write more variables; increase value of max_var by 50 in ../modeles/IOIPSL/src/restcom.f90
& max_var=550, max_file=50, max_dim=NF90_MAX_VAR_DIMS
then go and compile ORCHIDEE
on obelix
cd ../modeles/ORCHIDEE
First do a "normal" make to compile XIOS & IPOSL components which are needed; don't worry if the compilation of ORCHIDEE crashes
make
then compile
./makeorchidee_fcm -driver -arch ifort_LSCE -noxios
on curie
WARNING: before you do anything on curie; make sure you have copied the p86ipsl login environment to your home according to https://forge.ipsl.jussieu.fr/igcmg_doc/wiki/DocBenvBtgcc:
cp ~p86ipsl/.bashrc_curie ~/. cp ~p86ipsl/.bash_profile ~/.
Then compile the standard way:
cd ../config/ORCHIDEE_OL gmake without_xios
Optional: I usually re-compile ORCHIDEE again with makeorchidee_fcm to set options (don't forget to disable xios):
cd ../../modeles/ORCHIDEE ./makeorchidee_fcm -driver -parallel mpi -arch X64_CURIE -driver -noxios
4. Run ORCHIDEE (obelix)
4.1 SPINUP_ANALYTIC
4.1.1 create new experiment folder
cd ../../config/ORCHIDEE_OL/ cp -fr SPINUP_ANALYTIC/ MY_SPINUP_ANALYTIC/ cd MY_SPINUP_ANALYTIC/
4.1.2 tell libIGCM about the new input files (write me an email if you need them)
modify the variable "ListNonDel?" in COMP/stomate.card:
ListNonDel= (${R_IN}/SRF/reftemp.nc, .), \
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/USDA_SoilSuborder.nc, .), \
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/lithology.nc, .), \
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/deposition.nc, .)
WARNING: the deposition file is a static field at the moment; it does not include N deposition either. At the moment N deposition is set to P deposition in the ORCHIDEE code. Anyway N deposition is not accounted for in the calculations for the N & P cycle for now.
tell that we use the new “soil_param.nc”; in COMP/sechiba.card:
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/soils_param.nc, .), \
4.1.3 adjust the libIGCM
disable XIOS in COMP/orchidee_ol.card
# Use XIOS as output library instead of IOIPSL XIOS=n
disable XIOS in PARAM/run.def
# Use XIOS for writing diagnostics file # defulat = n XIOS_ORCHIDEE_OK = n
set the IO frequency to avoid crashes in PARAM/run.def
# Writefrequency in seconds in sechiba_history.nc # default = 86400.0 WRITE_STEP = 1800.0 # Writefrequency in days in stomate_history.nc # default = 10. STOMATE_HIST_DT = 1
add the new flags for the nutrient cycles in PARAM/run.def:
# to activate P cycle set =y STOMATE_OK_PCYCLE=y # to activate N cycle set =y STOMATE_OK_NCYCLE=y # make sure we dont fake LD_FAKE_HEIGHT=n # you can enable mass conservation and stoichiometry checks by =y STOMATE_MASS_CONSERVATION=y STOMATE_DSG_DEBUG=y
in addition you should add all the PFT and soil parameter values as set in /home/users/dgoll/ORCHIDEE/DEFS_n_CARDS/run.def
disable river routing if you run on non-global scale in run.def
# Activate river routing RIVER_ROUTING = n
4.1.4 adjust the job as usual
modify config.card as usual (see wiki)
modify run.def as usual (see wiki) plus if you want to archive, for example in the IMBALANCE-P storage; add to config.card:
ARCHIVE=/home/surface3/dgoll
4.1.5 create job
../../../libIGCM/ins_job
4.1.6 adjust job_file (optional)
Header to run a site scale simulation on obelix
###################### ## OBELIX LSCE ## ###################### #PBS -N test #PBS -m a #PBS -j oe #PBS -q short #PBS -o Script_Output_test.000001 #PBS -S /bin/ksh #PBS -v BATCH_NUM_PROC_TOT=1 #PBS -l nodes=1:ppn=1
to automatically have existing files deleted, set JobType?=DEV
#D- Experience type : DEB(ug), DEV(elopment), RUN (default) JobType=DEV
to avoid queuing take full advantage of your job time; the value depends on #PBS -q short and computation time of orchidee
#D- Number of execution in one job PeriodNb=60
to have a temp directory you can access on obelix; exchange dgoll with your ID
#D- Define running directory
#D- Default=${TMPDIR} ie temporary batch directory
#D-
#RUN_DIR_PATH=/workdir/or/scratchdir/of/this/machine
RUN_DIR_PATH=/home/scratch01/dgoll/RUN_DIR
4.1.7 submit the job
on obelix:
qsub Job_ID
on curie
ccc_msub Job_ID
4.2 ENSEMBLE: fluxnet sites http://forge.ipsl.jussieu.fr/orchidee/wiki/Scripts/FluxnetValidation
4.2.1 create new experiment folder
cd ../../config/ORCHIDEE_OL/ cp -fr ENSEMBLE/ MY_ENSEMBLE/ cd MY_ENSEMBLE/
4.2.2 tell libIGCM about the new input files (write me an email if you need them)
The configuration which are used are stored in ../SPINUP/SUBJOB/OOL_SEC_STO/COMP/ so, do as in 4.1.2: change the path of soil_param.nc and add the new files
sechiba.card:
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/soils_param.nc, .), \
stomate.card:
ListNonDel= (${R_IN}/SRF/reftemp.nc, .) \
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/USDA_SoilSuborder.nc, .), \
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/lithology.nc, .), \
(/home/scratch01/dgoll/ORCHIDEE/ORC-CNP/simulations/simple_forcing_DSG/deposition.nc, .)
WARNING: the deposition file is a static field at the moment; it does not include N deposition either. At the moment N deposition is set to P deposition in the ORCHIDEE code. Anyway N deposition is not accounted for in the calculations for the N & P cycle for now.
copy a run.def with all parameters needed to PARAM, for example this one:
cp ~/ORCHIDEE/DEFS_n_CARDS/run.def_NV PARAM/run.def
4.2.3 adjust the libIGCM
add the new flags for the nutrient cycles in fluxnet.card:
# to activate P cycle set =y STOMATE_OK_PCYCLE=y # to activate N cycle set =y STOMATE_OK_NCYCLE=y # make sure we dont fake LD_FAKE_HEIGHT=n # you can enable mass conservation and stoichiometry checks by =y STOMATE_MASS_CONSERVATION=y STOMATE_DSG_DEBUG=y
you need to add defaults for these switches into PARAM/orchidee.default
# STOMATE_OK_NCYLE ([FLAG]) : Activate the nitrogen cycle {OK_STOMATE}
STOMATE_OK_NCYCLE = n
# STOMATE_OK_PCYLE ([FLAG]) : Activate the phosphorus cycle {OK_STOMATE}
STOMATE_OK_PCYCLE = n
# STOMATE_MASS_CONSERVATION ([FLAG]) : activate mass conservation checks which force stop if violated {OK_STOMATE}
STOMATE_MASS_CONSERVATION = n
# STOMATE_DSG_DEBUG ([FLAG]) : activate checks for stoichiometry and negative pools {OK_STOMATE}
STOMATE_DSG_DEBUG = n
# LD_FAKE_HEIGHT ([FLAG]) : fakes height of vegetation (ORCHIDEE-CAN related) {OK_STOMATE}
LD_FAKE_HEIGHT = n
specify the spinup simulations in the config.card:
[SPINUP]
# Initialisation for spin-up :
# orchidee with sechiba alone (!!! if ok_stomate == n !!!)
duree_nostomate=0
# orchidee with stomate; number of years before n_iter cycles of duree_sechiba years of analytical spinup cycles
duree_inistomate=250
# teststomate (only if duree_inistomate > 0)
duree_offlineini=0
# Loop over ORCHIDEE runs (used for spin-up)
# The whole job is restarted n_iter times
# n_iter: the number of cycles with duree_sechiba years
n_iter=3
# orchidee with sechiba (and stomate if ok_stomate=y below)
duree_sechiba=50
# teststomate
duree_stomate=0
# forcesoil
duree_carbonsol=1
# Final run (full ORCHIDEE)
# This last parameter must be non-zero.
duree_final=100
xios is per default disabled in "PARAM/orchidee.default" routing is per default disabled in "fluxnet.card"
4.2.4 adjust the job as usual
modify config.card as usual (http://forge.ipsl.jussieu.fr/orchidee/wiki/Scripts/FluxnetValidation)
modify run.def as usual (http://forge.ipsl.jussieu.fr/orchidee/wiki/Scripts/FluxnetValidation) plus if you want to archive, for example in the IMBALANCE-P storage; add to config.card:
ARCHIVE=/home/surface3/dgoll
4.2.5 create job
../../../libIGCM/ins_job
4.2.6 adjust job (optional)
4.2.7 start simulations
my_prompt>> ksh my_prompt>> nohup ./Job_ENSEMBLE fluxnet > out_Job 2>&1 &
4.2.8 check simulations
ccc_mstat -u userid
ccc_mdel jobid
Attachments (11)
- missingPoints.png (20.5 KB) - added by dgoll 8 years ago.
-
USDA_map.png
(82.4 KB) -
added by dgoll 8 years ago.
The input file with the USDA soil orders in alphabetical order
-
USDA_map.2.png
(76.8 KB) -
added by dgoll 8 years ago.
input file with USDA soil orders in alphabetical order
-
USDA_map.3.png
(76.0 KB) -
added by dgoll 8 years ago.
The input file with the USDA soil orders in alphabetical order
-
lith_dom_map.png
(96.2 KB) -
added by dgoll 8 years ago.
dominant lithology
-
soilshield_map.png
(83.9 KB) -
added by dgoll 8 years ago.
soil shielding factor
-
ORC-CNP.png
(69.6 KB) -
added by dgoll 8 years ago.
Schematic representation of the key processes represented in ORCHIDEE-CNP
-
CNP_longterm.png
(46.6 KB) -
added by dgoll 8 years ago.
In red original formulation, in green the 3yr average. Shown is the C, the N and the CN of slow soil pool.
- BurnLabile.png (106.3 KB) - added by dgoll 8 years ago.
- Zoom.png (57.2 KB) - added by dgoll 8 years ago.
-
GPP_Hai.png
(48.2 KB) -
added by dgoll 8 years ago.
average annual cycle of DE-Hai site w & w/o N-limitation / w & w/o burning
Download all attachments as: .zip
